Simple TMVA regression
This blog entry will correspond to a simple TMVA regression using a Multi-Layer Perceptron (MLP) neural network and will entail details on how it was done. There are more things yet to be done (exciting!). The PTbin used for this is pt15_20 and can be found at the following directory-> /gpfs/mnt/gpfs01/star/data18/Run11JetEmbedding/JetTrees/pt15_20
The Toolkit for Multi-Variate Analysis (TMVA) comes with a tremendous amount of tools and different methods to try. TMVA accepts the simple TTree structure and in the case of this analysis, the Monte Carlo data came in jet-tree structure. The TTree that you will be using for TMVA must contain variables of the float type (Float_t) and all the variables must contain the same number of entries (as of now, that is the case. I'll have to look into it). Following were the steps taken to achieve this:
-
Grabbed the embedding file (embedding.cc) from Dr. Jim Drachenberg -> /star/u/drach09/Run11/Jets/gluonFF/embedding/embedding
-
Created two separate embedding files; embedding_fortmva_D.C & embedding_fortmva_P.C ->/star/u/rsalinas/embedding_fortmva_D.C & /star/u/rsalinas/embedding_fortmva_P.C
-
Using the TTree::AddFriend() function, I placed both trees in the "same tree" (done in the TMVARegression)
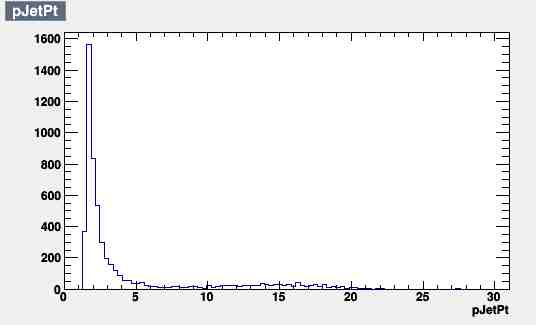
Each plot has 5379 entries

Error in <TTreeFormula::Compile>: Bad numerical expression :
> "myvariable"
> --- <FATAL> DataSetFactory : Expression myvariable could not
> be resolved to a valid formula.
> ***> abort program execution
Where "myvariable" is the variable you are trying to add to the factory. Once that is done you can go to /star/u/rsalinas and execute the following:
- make -f makefile.D
- make -f makfefile.C
- This creates the binaries necessary for the embedding
- ./RUN.sh
- This runs the embedding binaries for the particle and detector levels
- The output files should be called "outfilep.root" & "outfiled.root"
Once that is completed, you can run the TMVA regression file via the command:
- root -l TMVARegression.C\(\"MLP\"\)
- You can add more methods by separating them w/ commas \(\"LD,MLP,...\"\)
- Running this is the training aspect of the neural net.
TMVA has a nice GUI that displays how the training went and here are a few results:
- Re-scaled input variable and regression target
.jpg)
- Regression Output
- The weight per tree is 1
- 1000 training events and 4380 testing events
- Each neuron had an activation function of tanh
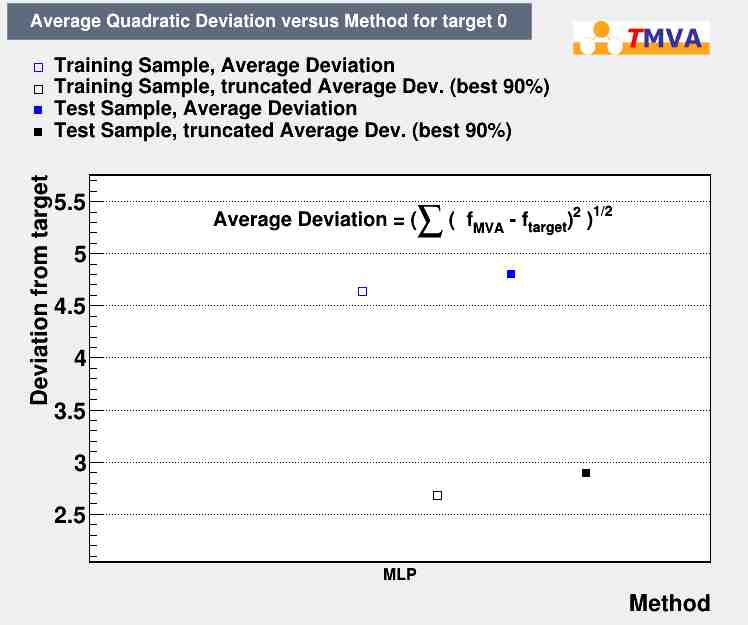
- From this simple example, you can see that there is little disagreement between the regression output and the true value used for testing

- The norm also shows small deviation
- The network architecture
- Two hidden layers consisting of 5 neurons each

While currently it seems to work well, this is a very simple example and will need further study and see how it compares with the unfolding package RooUnfold that is also native to ROOT. This method still yet has to be applied to actual data and will be given as an update to this blog entry.
- rsalinas's blog
- Login or register to post comments
