- dilks's home page
- Posts
- 2019
- 2018
- December (1)
- November (1)
- October (1)
- August (2)
- July (4)
- June (3)
- May (1)
- April (2)
- March (2)
- February (1)
- January (5)
- 2017
- December (3)
- November (1)
- October (2)
- September (3)
- August (2)
- July (2)
- June (1)
- May (2)
- March (3)
- February (3)
- January (3)
- 2016
- November (2)
- September (4)
- August (2)
- July (6)
- June (2)
- May (3)
- April (1)
- March (2)
- February (3)
- January (2)
- 2015
- December (3)
- October (3)
- September (2)
- August (6)
- June (3)
- May (3)
- April (4)
- March (3)
- February (5)
- January (3)
- 2014
- December (1)
- November (1)
- October (3)
- September (4)
- August (3)
- July (3)
- June (2)
- May (2)
- April (2)
- March (1)
- 2013
- 2012
- 2011
- My blog
- Post new blog entry
- All blogs
RP ADC and TAC Distributions
The following is data from the QT block read out of PP001, the RP QT boar.d for the 8 PMTs.
Below are ADC and TAC distributions as well as an inner vs. outer sequencer ("1" and "2" in RP group nomenclature) trigger overlap matrix. This is for day 85 pp transverse data.
.
.
.
.
.
RP Nomenclature
[ RP GROUP's NOMENCLATURE LINK ]
my nomenclature:
4 letters:
[East / West] [Inner / Outer] [Up / Down] [North / South]
.
.
Map check:
- I compared ADC thresholds output from my code to those from the JEVP plots and they all match. Features in ADC distributions also match, therefore I believe the map is correct.
- Furthermore, I compared the thresholds from day 66 data and day 85 data and they are the same. This indicates that ADC minimum cutoffs can be set the same for all runs.
- TAC distributions shift by up to ~50 counts between day 66 and 85
ADC min and TAC shift
ADC minimum cutoffs:
This is a zoom in of the MIP region of the full ADC distributions for each channel. The black line indicates the ADC minimum cutoff. The numbers are:
adcmin[0] = 110;
adcmin[1] = 110;
adcmin[2] = 110;
adcmin[3] = 90;
adcmin[4] = 120;
adcmin[5] = 90;
adcmin[6] = 110;
adcmin[7] = 110;
adcmin[8] = 120;
adcmin[9] = 115;
adcmin[10] = 120;
adcmin[11] = 120;
adcmin[12] = 130;
adcmin[13] = 90;
adcmin[14] = 125;
adcmin[15] = 100;
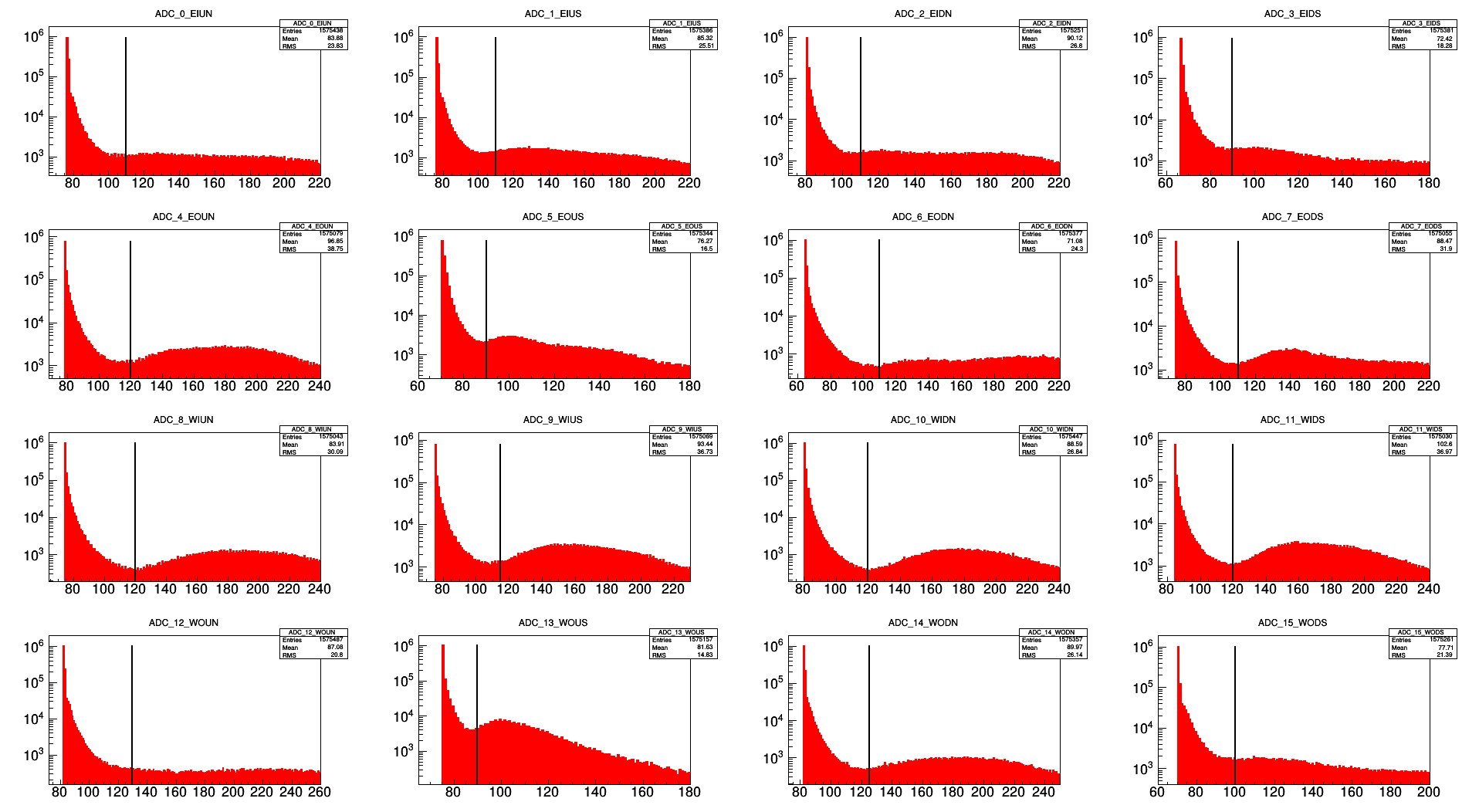
.
.
TAC peak shifts:
The TAC counts are subtracted such that the maximum bin is at 1000 counts. The image below shows the TAC distrubions before the subraction. The numbers are:
TACcalib[0] = 553;
TACcalib[1] = 537;
TACcalib[2] = 528;
TACcalib[3] = 600;
TACcalib[4] = 628;
TACcalib[5] = 446;
TACcalib[6] = 491;
TACcalib[7] = 521;
TACcalib[8] = 338;
TACcalib[9] = 290;
TACcalib[10] = 440;
TACcalib[11] = 163;
TACcalib[12] = 704;
TACcalib[13] = 768;
TACcalib[14] = 642;
TACcalib[15] = 757;
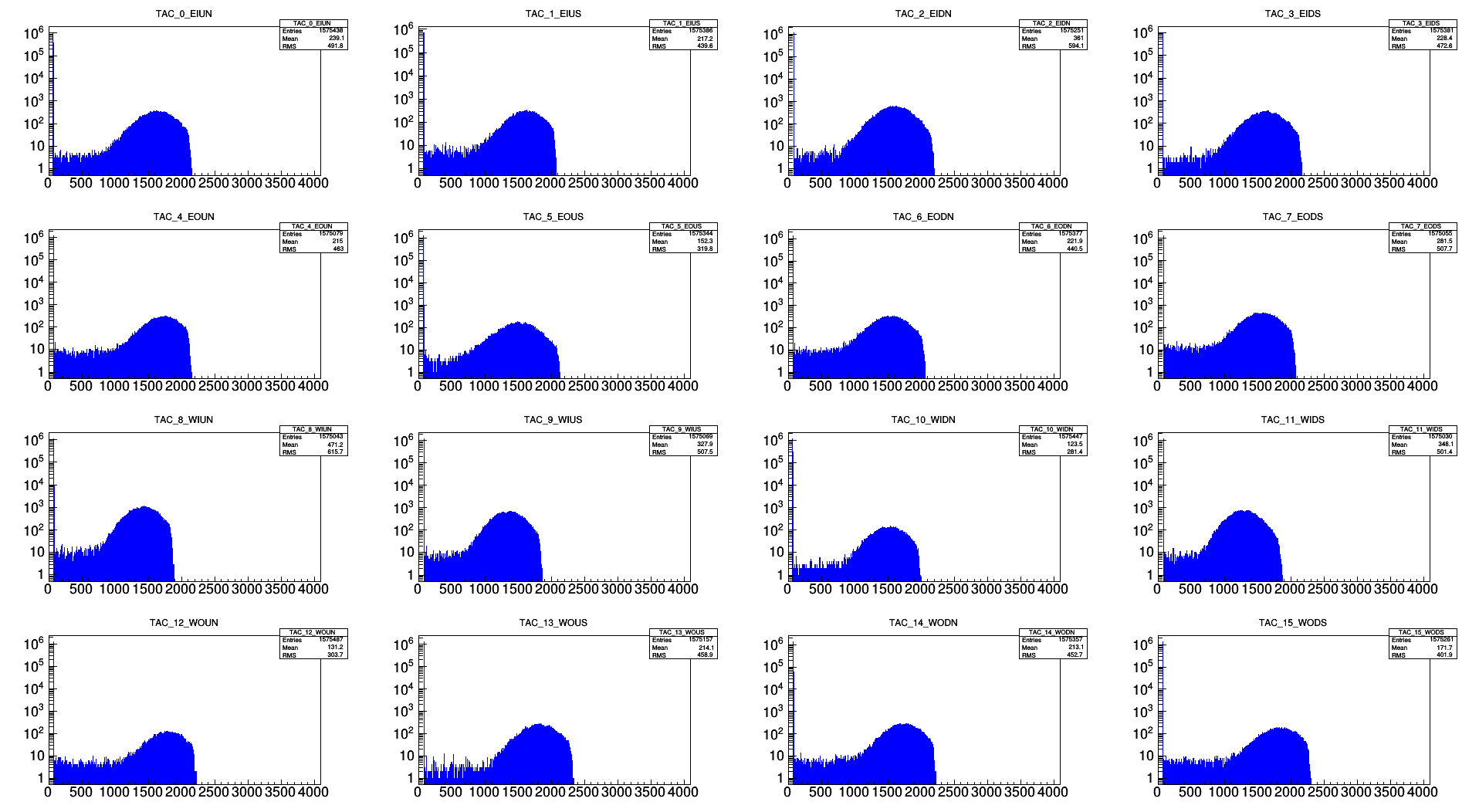
.
.
.
.
Day 85 Full ADC Distributions
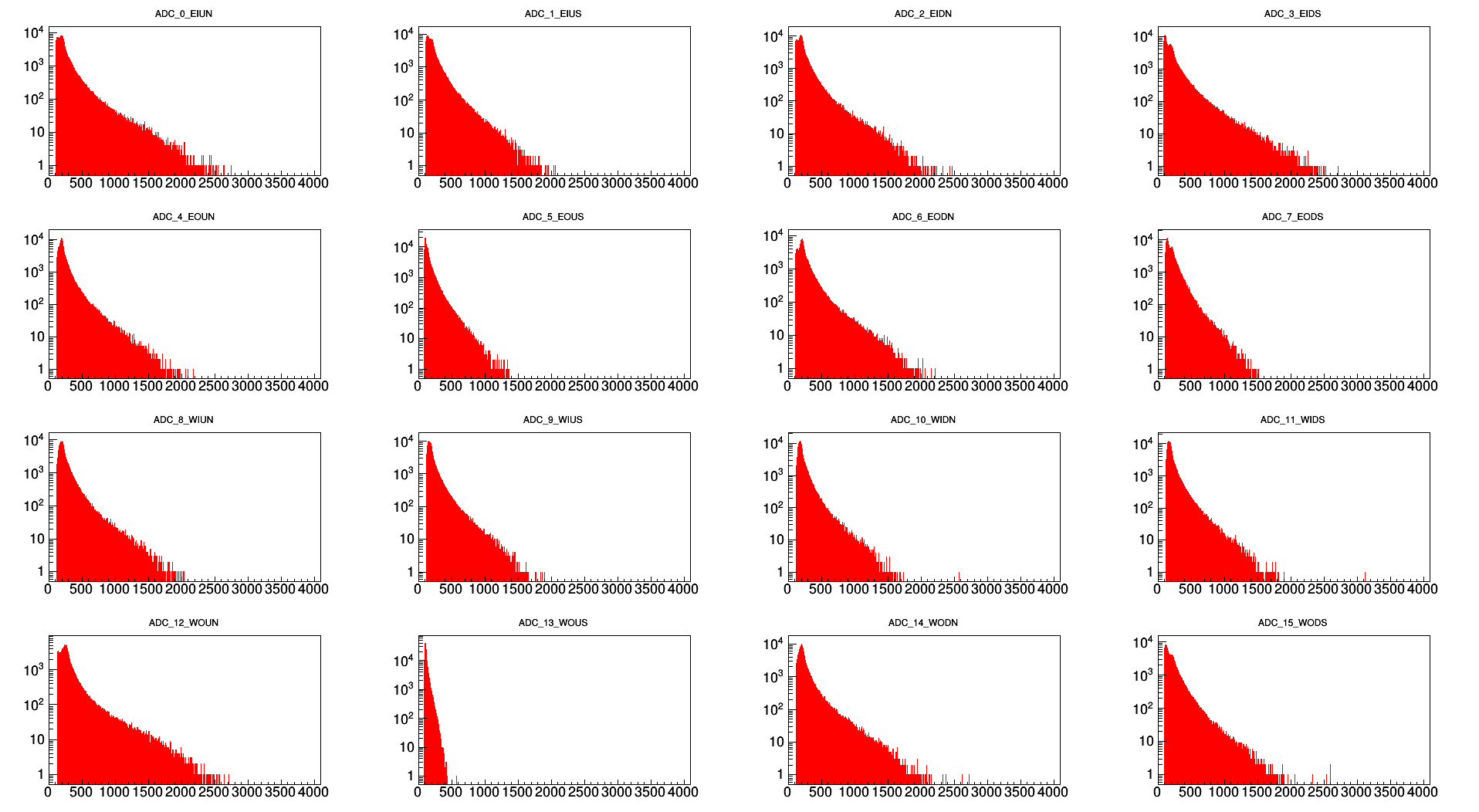
.
.
.
.
Day 85 Shifted TAC Distributions
.png)
.
.
.
.
Day 85 Multiplicty, Vertex, and Inner/Outer Sequencer Overlap
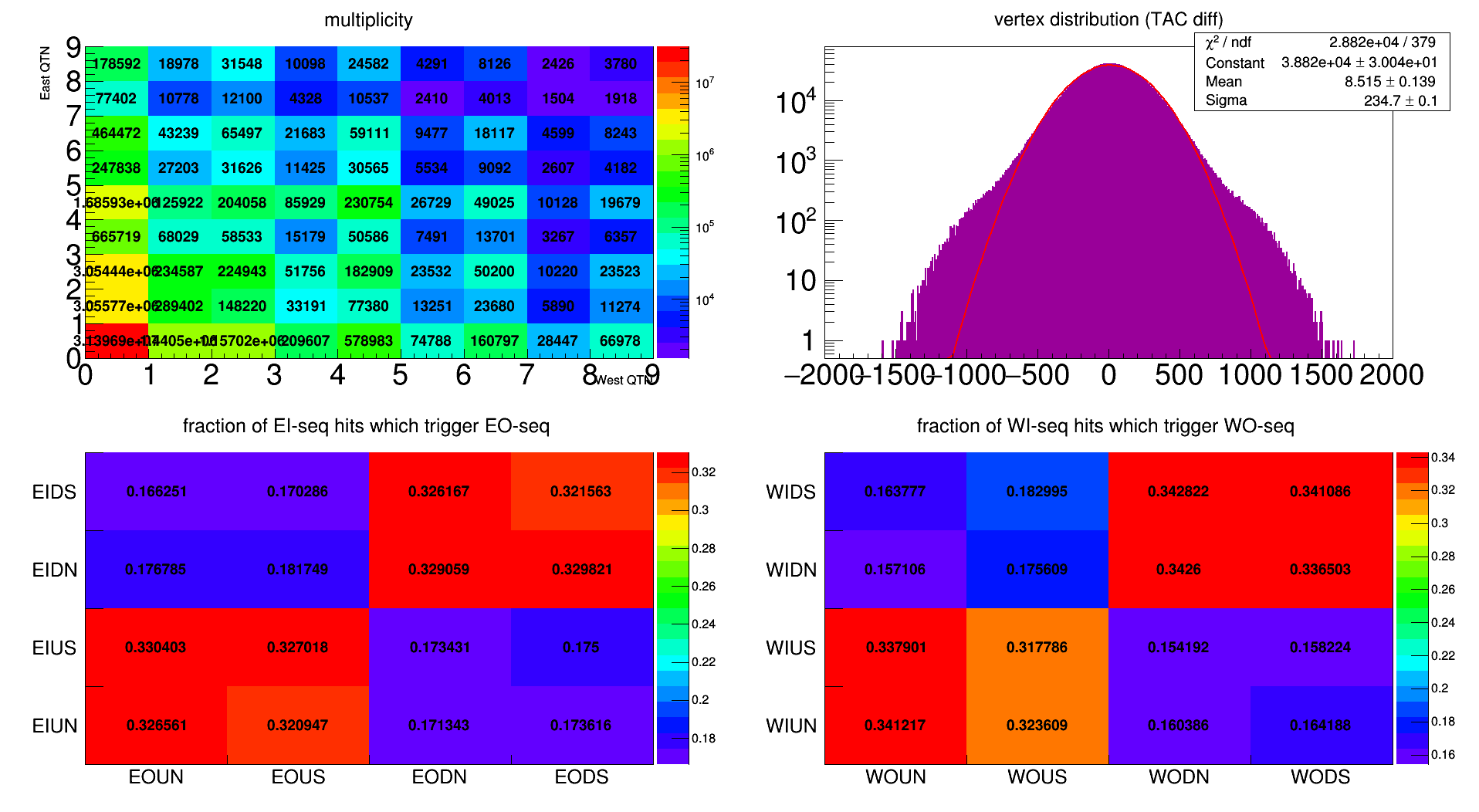
- dilks's blog
- Login or register to post comments
