computing
ITD's Catalyst chassis in the DAQ room (subnets 60, 162, wireless and possibly others in 1006)
This switch chassis is in the networking rack in the northwest corner of the DAQ room. It is managed by ITD. STAR has no way to interact with this switch at the software/confi
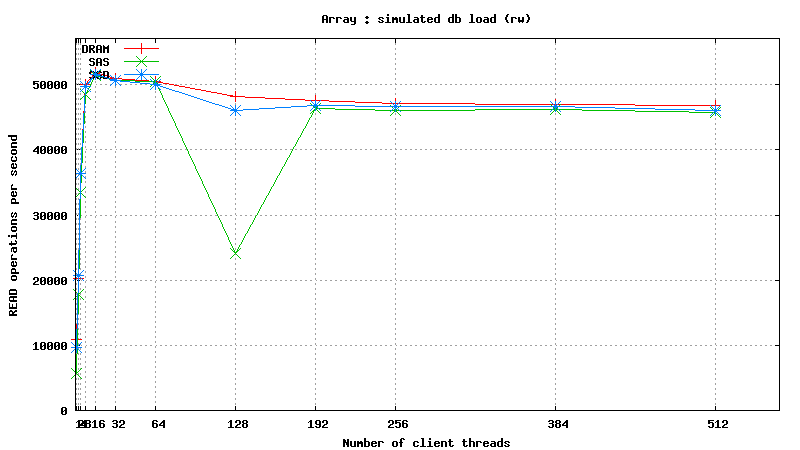
Simulated DB Load : DRAM vs SSD vs SAS
Simulated DB Load : SSD
Simulated DB Load : Solid State Disk
SysBench parameters: table with 20M rows, readonly. Allowed RAM limit: 2Gb to reduce fs caching effects.
Simulated DB Load : SAS
Simulated DB Load : Serial Attached SCSI
SysBench parameters: table with 20M rows, readonly. Allowed RAM limit: 2Gb to reduce fs caching effects.
Simulated DB Load : DRAM
Simulated DB Load Testing Results : DRAM
SysBench parameters: table with 20M rows, readonly. No RAM limit, /dev/shm was used as MySQL data files location.
Rules to Live By in Online Networking at STAR
Updated on Fri, 2010-07-09 13:51. Originally created by wbetts on 2010-06-18 14:50. Under:Document everything!
All hardware with an IP address should be labelled.
production-status-update-run-2010
Updated on Mon, 2011-06-13 08:27. Originally created by didenko on 2010-06-07 09:24. Under:Run 2010 AuAu 7.7GeV - 200GeV data production
Last update on 2011-06-13
Production Status for run 2010
Updated on Mon, 2010-06-07 16:14. Originally created by didenko on 2010-06-07 09:11. Under:For EC and ED(s) (OBSOLETE)
Embedding instructions for EC and ED(s)
Obsolete documentations
Updated on Thu, 2010-05-20 19:28. Originally created by starembed on 2010-05-20 19:26.
Under:
For Embedding Helpers (OBSOLETE)
Embedding instructions for Embedding Helpers
Dynamic Random Access Memory testing results
SysBench results for tmpfs using 6 GB of DRAM
[ SAMSUNG DDR3-1333 MHz (0.8 ns) DIMM, part number : M393B5673DZ1-CH9 ]
Read more
Solid State Drive testing results
SysBench results for Intel® X25-E Extreme SATA Solid-State Drive
Serial Attached SCSI drive testing results
SysBench results for Fujitsu MBC2073RC
Solid State Drives (SSD) vs. Serial Attached SCSI (SAS) vs. Dynamic Random Access Memory (DRAM)
INTRODUCTION
This page will provide summary of SSD vs SAS vs DRAM testing using SysBench tool. Results are grouped like this :
CPU and bulk storage purchase 2010
TPC de/dx
The TPC dE/dx calibration documentation from Run 11 can be found at the following:
http://drupal.star.bnl.gov/STAR/blog/yiguo/2012/feb/01/runxi-dedx-calibration-recipe
201004
The topics were a "classic":
- Action items since last group meeting + team matters
- Focus activities
- Q&A
Obsolete documentations
Updated on Thu, 2010-05-20 19:28. Originally created by starembed on 2010-05-20 19:26. Under:For Embedding Helpers (OBSOLETE)
Embedding instructions for Embedding Helpers
Dynamic Random Access Memory testing results
SysBench results for tmpfs using 6 GB of DRAM
[ SAMSUNG DDR3-1333 MHz (0.8 ns) DIMM, part number : M393B5673DZ1-CH9 ]
Solid State Drive testing results
SysBench results for Intel® X25-E Extreme SATA Solid-State Drive
Serial Attached SCSI drive testing results
SysBench results for Fujitsu MBC2073RC
Solid State Drives (SSD) vs. Serial Attached SCSI (SAS) vs. Dynamic Random Access Memory (DRAM)
INTRODUCTION
This page will provide summary of SSD vs SAS vs DRAM testing using SysBench tool. Results are grouped like this :
CPU and bulk storage purchase 2010
TPC de/dx
The TPC dE/dx calibration documentation from Run 11 can be found at the following:
http://drupal.star.bnl.gov/STAR/blog/yiguo/2012/feb/01/runxi-dedx-calibration-recipe
201004
The topics were a "classic":
- Action items since last group meeting + team matters
- Focus activities
- Q&A