BeamLine Constraint
Future suggestion: determine beamline width via impact parameter correlations
Beamline Calibration How-To
This is a step-by-step outline of how to perform the beamline calibrations. Results from each year of running can be found in the child page links below.
- Decide upon usable triggers.
- Preferrably minbias triggers, though some special triggers are fine too. The point is that the trigger should not have a bias on the reconstructed primary vertex positions (an event-wise bias is acceptable, as long as the average over many events yields no overall bias). A trigger that selectively prefers events at, for example, positive z versus negative z, is not a bias which concerns us. Meanwhile, a trigger which selects triggers using tracks from only one patch in phi in the TPC may indeed bias the vertex-finding because of track energy loss, or SpaceCharge, or other distortions.
Deciding upon these triggers may involve making a few calibration passes on a subset of the data with enough statistics to see different biases. So if there is any uncertainty about the triggers to use, there may be some iteration of the following steps.
There is also a triggers webpage which provides helpul information about the various triggers (NOTE: its URL changes for each year, so try similar URLs or parent directories for different years). - Decide upon filetype to use.
- If there are files already available with all the other calibrations (TPC alignment, T0, drift velocity, twist, etc.) in place, then one can process data faster by using either event.root (fast) or MuDst.root (really fast) files. As of this writing, one can process either of these using the FindVtxSeeds.C macro:
void FindVtxSeeds(const Int_t mode=0, const Int_t nevents=10, const Char_t *path="/star/data13/reco/dev/2001/10/", const Char_t *file="st_physics_2304060_raw_0303.event.root", const Char_t* outDir="./");where mode is unused, and the other parameters are self-explanatory. In this case, output files go to the outDir. The output files will be discussed later.If no available DST files will suffice, then the DAQ files will need to be processed using BFC options including "VtxSeedCalG", plus the year's geometry, a tracker, a vertex finder, and possibly other correction options. For example, in 2008 we used "VtxSeedCalG,B2008,ITTF,VFMinuit,OSpaceZ2,OGridLeak3D". The output files will, by default, go into either the local directory, or ./StarDb/Calibrations/rhic/ if it exists.
- Choosing the actual files to use.
- Now it is time to query the database for the files to use. My approach has been to link several DB tables which provide a lot of information about fill numbers, triggers, numbers of events, etc, in an effort to somewhat randomly choose files such approximately 10,000 useful triggers are processed from each fill. To do this, I passed queryNevtsPerRun.txt to mysql via the command:
cat query.txt | mysql -h onldb.starp.bnl.gov --port=3501 -C RunLog > output.txt
where the query file must be customized to select the unbiased triggers, triggersetups, and runs/fills. The query has been modified and improved in recent years to avoid joining several very large tables together and running out of memory for the query - see for example this query. This now looks much more complicated than it really is. Several simple cuts are placed on trigger setups, trigger ids (by name), run sanity (status and non-zero magnetic field), inclusion of the TPC, file streams (filenames), and lastly the number of events in the files and some very rough math to select some number of events per run by using some fraction of the file sequences.This should return something like 10,000 events per run accepted by the query. The macro StRoot/macros/calib/TrimBeamLineFiles.C can be used to pare this down to a reasonable number of events per fill (see the macro for more detailed instructions on its use). This macro's output list of files then can be processed using the chain or macro as described above.
- The output of the first pass.
- The output of the VtxSeedCal chain or FindVtxSeeds.C macro consists of anywhere from zero to two files. The first, and most likely, file will be something like: vertexseedhist.20050525.075625.ROOT (I give the output filename a capital suffix .ROOT to avoid it being grabbed by the DB maker code when many jobs used the same input directory, and it should be renamed/copied/linked such that its name has a lower case suffix .root for subsequent work). This file will appear if at least one usable vertex was found, and it contains an ntuple of information about the usable vertices. The timedate stamp will be that of the first event in the file, minus a few seconds to insure its validity for the first event in the file. The second possible file will be of the form vertexSeed.20050524.211007.C and contains the results of the beamline fit, which is performed only if at least 100 usable vertices are found. These table files are only useful as a form of quick QA, but are not needed for the next steps.
- Decide on cuts to remove biases and/or pile-up
- Use the ntuple files to look at vertex positions versus various quantities to see if cuts are necessary to remove biases and/or pile-up. Documentation on what can be found in the ntuples is available in the StVertexSeedMaker documentation. For example, one may open several ntuple files together in a TChain and then plot something like:
resNtuple.Draw("x:mult>>histo(10,4.5,54.5)","abs(z)<10");...to see if there is a low-multiplicity contribution from pile-up collisions (note that the above is but one example of many possible crosschecks which can be made to look for pile-up). Any cuts determined here can be used in the next step. - The second (aggregation) pass.
- All the desired ntuple files from the first pass should be placed in one directory. It is then a simple chore of running the macro (which is in the STAR code library and will atomatically be found):
root4star -b -q 'AggregateVtxSeed.C(...)' Int_t AggregateVtxSeed(char* dir=0, const char* cuts="");
where dir is the name of the directory with the ntuple files, and cuts is any selection criteria string (determined in the previous step) formatted as it would be when using TTree::Draw(). The output of this pass will be just like that of the first pass, but will reduce to one file per fill. Again, no ntuple file without at least one usable vertex, and no table file without at least 100 usable vertices. Timedate stamps will now represent the start time of fills. These output table files are what go into the DB after they have been QA'ed. - QA the tables.
- I like to look at the table values as a function of time graphically to do QA. I used to do this with shell scripts, but now the aggregation pass should output a file called BLpars.root which summarizes the calibration results. The data in that file can easily be plotted by running the another macro, which is also in the STAR code library and will automatically be found):
root 'PlotVtxSeed.C(...)' BLpars->Draw("x0:zdc");The intercepts and slopes will be plotted on one graph (see the 2006 calibration for some nice examples of the plots), and a TTree of the results will be available for further studies using the pointer BLpars. You might also be interested to know that this macro can work on a directory of vertexseedhist.*.root files as the BLpars ntuples are stored in them as well (alongside the resNtuple ntuples of individual vertices, but only if a vertexSeed.*.C file was written [i.e. if BeamLine parameters were found]) and can be combined in a TChain, which the macro does for you.
- Here are some example slopes and intercepts from my first look at QA on 2005 data (note that these plots were made from older scripts, not PlotVtxSeed.C):
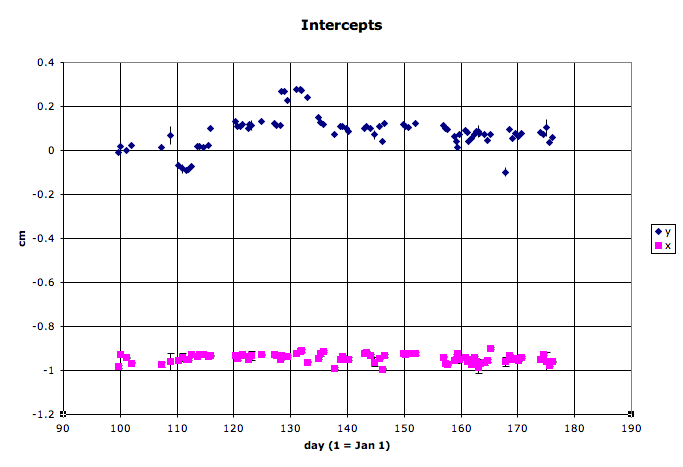
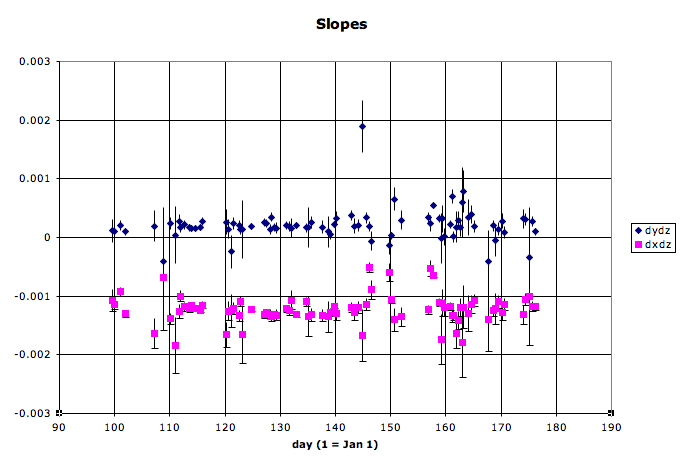
One can see several outliers in the data in addition to the normal trends of machine operations. Many of these outliers are due to limited statistics for a fill. Care must be used in deciding what to do:- If the statistics are great for that fill, this may be a real one-fill alteration of beam operations by RHIC, and the datapoint should be kept.
- If there is a long gap in data before that point, then it may be wise to keep the datapoint.
- If statistics are poor (usually the case with about 1000 or fewer usable vertices from a fill), and the data on either side of the point appears stable, it is probably a good idea to remove the datapoint.
- It may be possible to run some additional files from that fill to increase the statistics to the level where the datapoint is more significant.
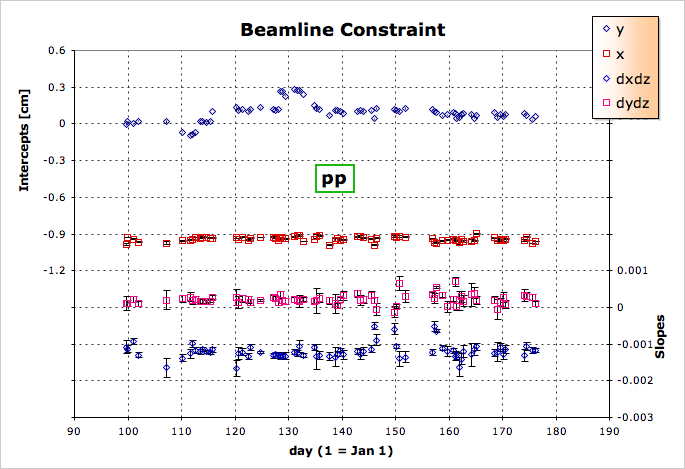
- Enter the tables into the DB.
- You will need to have write permission to the Calibrations_rhic/vertexSeed table. If you do not, please see the STAR Calibrations Coordinator to arrange this with the STAR Database Administrator (see the Organization page). The DB admin can also give you examples on how to upload tables. But I will include here an example using a macro of mine which is obtainable via CVS from offline/users/genevb/WriteDbTable.C. Place all the table files into one directory from which you will upload them to the DB. I use ~/public/vertexSeed as that directory in the example below:
setenv DB_ACCESS_MODE write root4star -b 'WriteDbTable.C("~/public/vertexSeed","vertexSeed","vertexSeed","Calibrations_rhic")'The number of successfully (and unsuccessfully) loaded tables should be listed. - ...and you are done.
- Printer-friendly version
- Login or register to post comments
